Milestone 2: Incorporating dna sequence in SBOL file
-
Started exploring the dna sequencing part
-
Explored the components used in the circuit
-
What is RBS? CDS? Promotor? Terminator? Ribozyme? where are they placed
-
How does SBOL format work
-
How is the current code doing it. What changes are needed?
-
Had power cuts and internet issues. So hard to maintain the same productivity. Progress got a bit slow
- CELLO:
- studied main paper (not entirely because not familiar with the technicalities, mainly the tables and figures) https://science.sciencemag.org/content/352/6281/aac7341
- studied supplementary material (not entirely because not familiar with the technicalities, mainly the tables and figures) https://science.sciencemag.org/content/sci/suppl/2016/03/30/352.6281.aac7341.DC1/Nielsen.SM.pdf
- Their git repo. SBOL .xml examples to see how the dna sequencing is embedded in it
- Their website to see what they generate and how they do it
- Compared their circuit .xml file to our sample .xml files. Figured out:
- They mostly have insulated circuits.
- We are making non-insulated gate circuits
- GeneTech doesn’t specify what terminator it is using (we just use a dummy id for the terminator)
- GeneTech has no information of the RBS whatsoever (its not even included)
- For non-insualted circuits, Cello paper had the same terminator
- SBOL:
- read briefly the data specification document (https://sbolstandard.org/wp-content/uploads/2016/06/SBOL2.3.0.pdf)
- GeneTech report by Bhutto et. al to understand how they are doing it
- pySBOL documentation
- pySBOL paper
- Created an example device to see how it works
- sbol tutorials:
- https://sbolstandard.org/wp-content/uploads/2016/08/pysbol-crispr-tutorial.pdf
- tutorial on their github
- exampels on their github.
- Made a database of the parts used in our Circuits:
- All Input promoters
- All output promoters
- All CDS
- Included a terminator
- YFP
- The links to the repository for each of these
- Used pysbol to query the link for each of these and populated the database.
- This is how our database looks now: See here
-
Exams break from 1st July to 12th July, some progress here and there (included in the rest below).
-
As for the two issues that we were facing, the first one was that of a terminator. I sticked to a single terminator for all terminators.
-
The terminator and it’s sequence as well as it’s repository url is included in the database
-
As for the choice of RBS, we had initially agreed to use a dummy string for the RBS since for non-insulated circuits we are not aware of how to choose RBS
-
I initially did this but then thought of incorporating RBS as well, and to that end, used the first RBS corresponding to a gate given in the Nielson paper
-
P1 for PhlF, H1 for HlYllR, A1 for AmeR, B1 for Betl and so on. This information again is highlighted in the in the database
-
I did this so that in future we could easily make a small change and we would be able to incoporate more robust RBS. I created a gate to RBS mapping in the code for this.
-
-
Alignment of parts into a final sequence
- For this part I studied the Nielson paper and supplemtary material, they mentioned that this is done linearly
however due to the lack of examples, I wasn’t able to fully grasp how to do that. I looked into additional material
but couldn’t figure out how to do it. Then finally I downloaded a bunch of SBOL files corresponding to the circuits
given in the paper, and saw how they were doing it. As an example, find this circuit below:
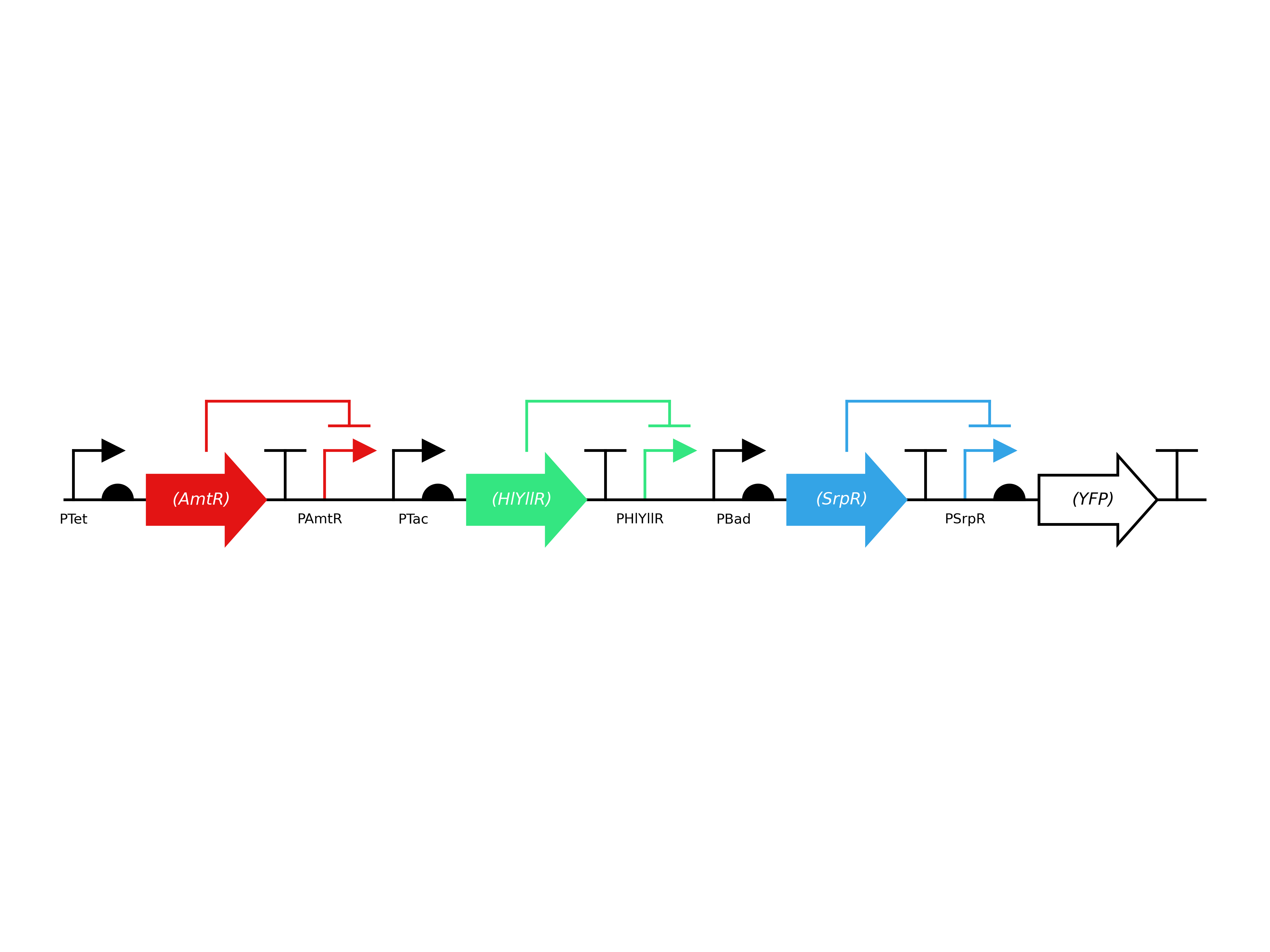 To get the final sequence of the circuit, the components are aligned in the following linear order:
To get the final sequence of the circuit, the components are aligned in the following linear order:
-
PTet, RBS for AmtR, AmtR, Terminator, PAmtR, …..
-
you might have noticed that the dna sequence of components appears in the final sequence in the same order as it appears in the sbol visual circuit.
-
- Furthermore, each part’s sequence was added the sbol file using the Sequence() class in pysbol and it was linked to it’s corresponding component definition. It can be seen in the following image:
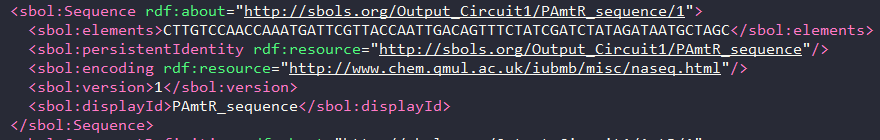
-
Each component definition was then used to compile the dna sequence of the final circuit.
-
Note that we use the same terminaor at all terminator locations. This is replicated in the final sequence as well
-
A sequence annotation object is also created for all components highlighting where the dna sequence of that component is placed in the final sequence as follows:
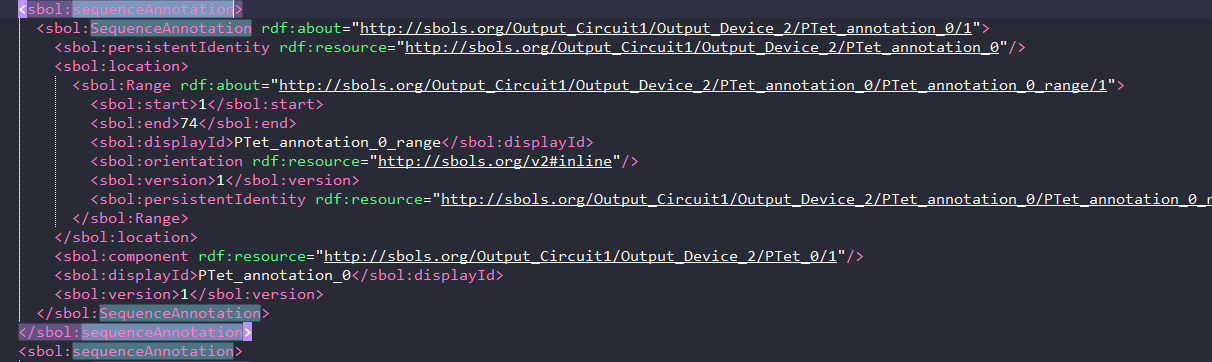
- For this part I studied the Nielson paper and supplemtary material, they mentioned that this is done linearly
however due to the lack of examples, I wasn’t able to fully grasp how to do that. I looked into additional material
but couldn’t figure out how to do it. Then finally I downloaded a bunch of SBOL files corresponding to the circuits
given in the paper, and saw how they were doing it. As an example, find this circuit below:
- There were a few bugs in the existing code. I tried to fix them as I encountered them. One bug that particularly bothered me was that in the SBOL file, in one of the loops instead of using the loop variable, the code used a constant number.
- componentDef_string[j][1] instead of componentDef_string[j][k].
- This took a lot of digging and wasted a lot of time. But it worked at the end.
-
There are some bugs that appear randomly in functions.py (written by previous contributors). I haven’t been able to trace those yet
-
Second milestone is now complete!
-
I will open a pull-request for this on the main repository shortly.
- Cleaned the repository.
Reference
- https://synbiohub.programmingbiology.org/public/Eco1C1G1T1/YFP/1 (Info of YFP Sequence SynBioHub)
- https://pysbol2.readthedocs.io/en/latest/getting_started.html (pysbol2)
- https://sbolstandard.org/wp-content/uploads/2015/07/pySBOL-tutorial.pdf (A brief description of the CRISPR circuit using SBOL 2.0 data model)
- https://readthedocs.org/projects/pysbol2/downloads/pdf/latest/ (pysbol reference guide)
- GeneTech2.0 Developers’ Guide
- GeneTech2.0 Report
- https://science.sciencemag.org/content/sci/suppl/2016/03/30/352.6281.aac7341.DC1/Nielsen.SM.pdf (Nielson’s Supplementary Material)
- https://science.sciencemag.org/content/352/6281/aac7341 (Genetic Circuit Design Automation)
- https://sbolstandard.org/wp-content/uploads/2016/06/SBOL2.3.0.pdf (SBOL reference guide)
- https://github.com/CIDARLAB/cello/(Cello’s repository)
